Water buffalo (Bubalus bubalis) is an important source of meat and milk in countries with relatively warm weather. Compared to the cattle genome, a little is done to reveal the genome structure of the buffalo. Production of a high resolution draft for whole genome of Egyptian buffalo (Bubalus bubalis) will increase the role of genome in the different disciplines of livestock research and promote agricultural sustainability for Egypt.
We introduce a high quality draft assembly of the Egyptian buffalo genome, as a representative of the water buffalo species. The sequencing was based on the Next Generation Sequencing technology. The assembly was achieved using a reference-based assembly workflow, which dramatically reduces the computational complexity of the assembly process and integrates different public resources to improve the assembly quality.
Identification of genetic features within the sequenced genomes, such as genes, repeats, etc, which enable better understanding of the animal traits, physiology, domestication and evolution of the buffalo species. We also introduce further analysis, including genome annotation, variations among different animals from the same species, and comparison to the mammalian genomes.

The sequencing was based on the 5500xl SOLiD sequencer, which is a next generation sequencing technology. We achieved about 50-60x coverage of the genome, to enable good sequencing results. We used different types of libraries to produce two sets of reads

The workflow is based on using a reference genome, which is the cattle in our case, to sequence the target genome. The use of the reference genome is intended to speed up the assembly process without comprising the quality of the assembly.

To capture the contiguous parts of the Buffalo genome, The algorithm works by combining reads mapped to the reference genome and de-novo assembly to create the contigs from the reads.
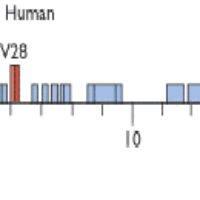
To spot repetitive parts of the genome and categroize them into different repeat families.
The scaffolding process runs in a multistep manner using mate-paired reads along with evidences coming from the reference genome to reduce the complexity of the scaffolding problem.
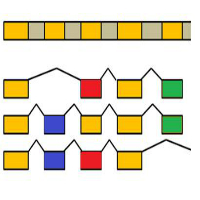
Genome annotation of the assembled genome involves the determination of genes with their exon-intron structure.
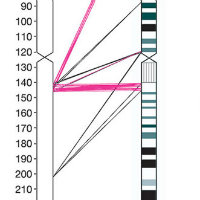
Comparison to genomes of other species and identifying rearrangement events
Here links to download runs, contigs, and scaffolds data



The Center and the research project is funded by the Science and Technology Development Fund (STDF), Egypt.
Our analysis pipeline is optimized to speed up the analysis. It also combines information from public resources to improve the assembly. We used a number of tools including velvet, Bowtie, SHRiMP, BLAST, BLAT in a complex pipeline to finish the assembly and annotation. Go the Tavaxy Workflow